Whole Exome Sequencing
Whole Exome Sequencing (WES) is a high-throughput, next generation sequencing-based assay analyzing the whole human exome of approximately 23,000+ DNA genes. Caris’ testing detects DNA Point Mutations Single Nucleotide Variants (SNVs), Insertions and Deletions (InDels), Copy Number Alterations (CNAs), Karyotyping, Viruses, Homologous Recombination Deficiency (HRD), Genomic Loss of Heterozygosity (gLOH), Microsatellite Instability (MSI), Tumor Mutational Burden (TMB) and Human Leukocyte Antigen (HLA) genotyping, all from one streamlined test.
Whole Exome Sequencing (WES) DNA
- 23,000+ genes
- SNVs, inDels, copy number alterations & karyotyping
- Evenly-spaced genomic SNP
- Genomic signatures:
- Other:
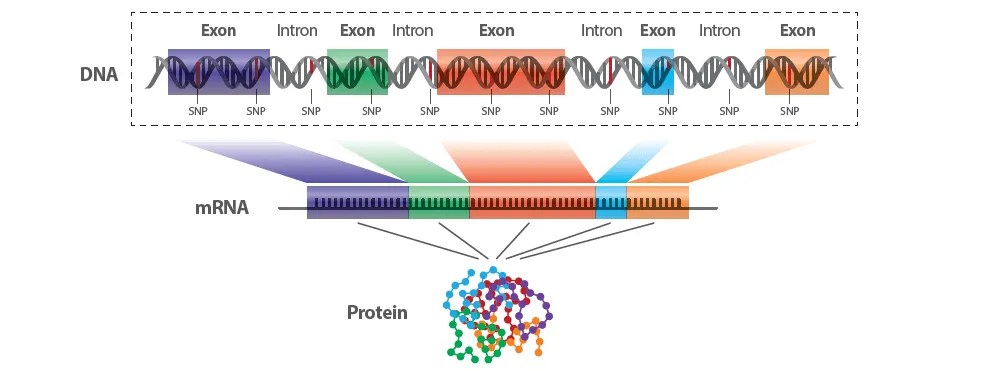
Exons are 1-2% of the genome that encode the proteins
Exome = Total exons of the genome
Example Gene
Discover MI Cancer Seek®, the first and only WES/WTS-based assay with CDx indications for both adult and pediatric patients.
Complete Gene Coverage with Caris Molecular Testing
As the pioneer in precision medicine, Caris was the first to provide comprehensive Whole Exome and Whole Transcriptome sequencing for every patient. Each Caris molecular profiling order includes next-generation sequencing of all 23,000+ genes.
Looking for a specific gene? Use the full gene search below to verify that it is included in Caris profiling, or browse the list of genes most commonly associated with cancer.
Discover
More
Caris’ Whole Transcriptome Sequencing (WTS) uses the capabilities of high-throughput sequencing to gain insight into the RNA profiles of patient tumors.
Genomic Loss of Heterozygosity or genomic instability is often related to defective homologous recombination repair mechanisms.
